Copyright © 2007-2010 BiochemLabSolutions.com
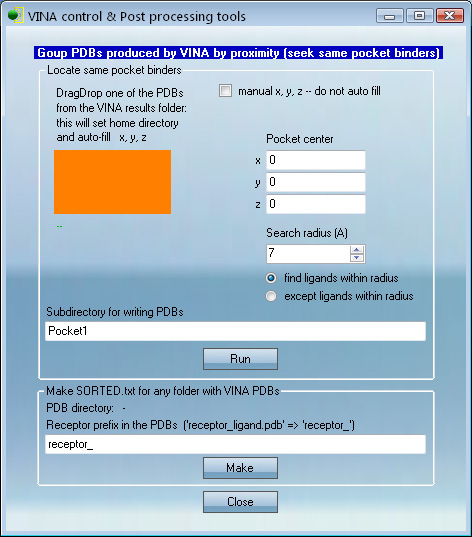
VcPpt contains tools for finding PDBs in the screening run that are within a certain distance from a chosen ligand or a pocket.

If you have a ligand and need to find which other ligands were docked nearby:
1. Drag-Drop this ligand's PDB to the orange pad; x, y and z will be filled pointing at the
Drag-Dropped ligand's center.
PDBs for searching must be present in the same folder with this ligand.
2. The "RUN" button will find PDBs whose top scoring solution is within 7A (distance is adjustable)
from the x, y, z. These PDBs will be copied to the subfolder "Pocket1".
- x, y, z can be entered manually to specify any position on a protein surface
- x, y, z can also be computed by copying a few amino acids from the pocket of interest in target protein to a PDB file and Drag-Dropping this file to the orange pad to set x, y, and z of the pocket.
"Manual x, y, z -- do not auto fill" should next be checked to fix the coordinates. Then Steps 1 and 2 above are used.
Any manual x, y, z can also be set.
_____________________
All sorting is done based on the top docked solution in the PDB files. By combining the sorting by scores and sorting by binding position it takes minutes to find the best binders to any given pocket among thousands of docking results.
1. Drag-Drop this ligand's PDB to the orange pad; x, y and z will be filled pointing at the
Drag-Dropped ligand's center.
PDBs for searching must be present in the same folder with this ligand.
2. The "RUN" button will find PDBs whose top scoring solution is within 7A (distance is adjustable)
from the x, y, z. These PDBs will be copied to the subfolder "Pocket1".
- x, y, z can be entered manually to specify any position on a protein surface
- x, y, z can also be computed by copying a few amino acids from the pocket of interest in target protein to a PDB file and Drag-Dropping this file to the orange pad to set x, y, and z of the pocket.
"Manual x, y, z -- do not auto fill" should next be checked to fix the coordinates. Then Steps 1 and 2 above are used.
Any manual x, y, z can also be set.
_____________________
All sorting is done based on the top docked solution in the PDB files. By combining the sorting by scores and sorting by binding position it takes minutes to find the best binders to any given pocket among thousands of docking results.

Find ligands that cluster in 3D space